Domesticated 6,000–10,000 years ago in the Andean highlands of southern Peru [1, 2], the cultivated potato (Solanum tuberosum L.) has quickly become one of the major staple foods worldwide [3]. With a global production of 377 million tonnes in 2016, for a total gross value of $111 billion, the potato is now the fourth most important food crop in the world, after rice, wheat and corn [4].
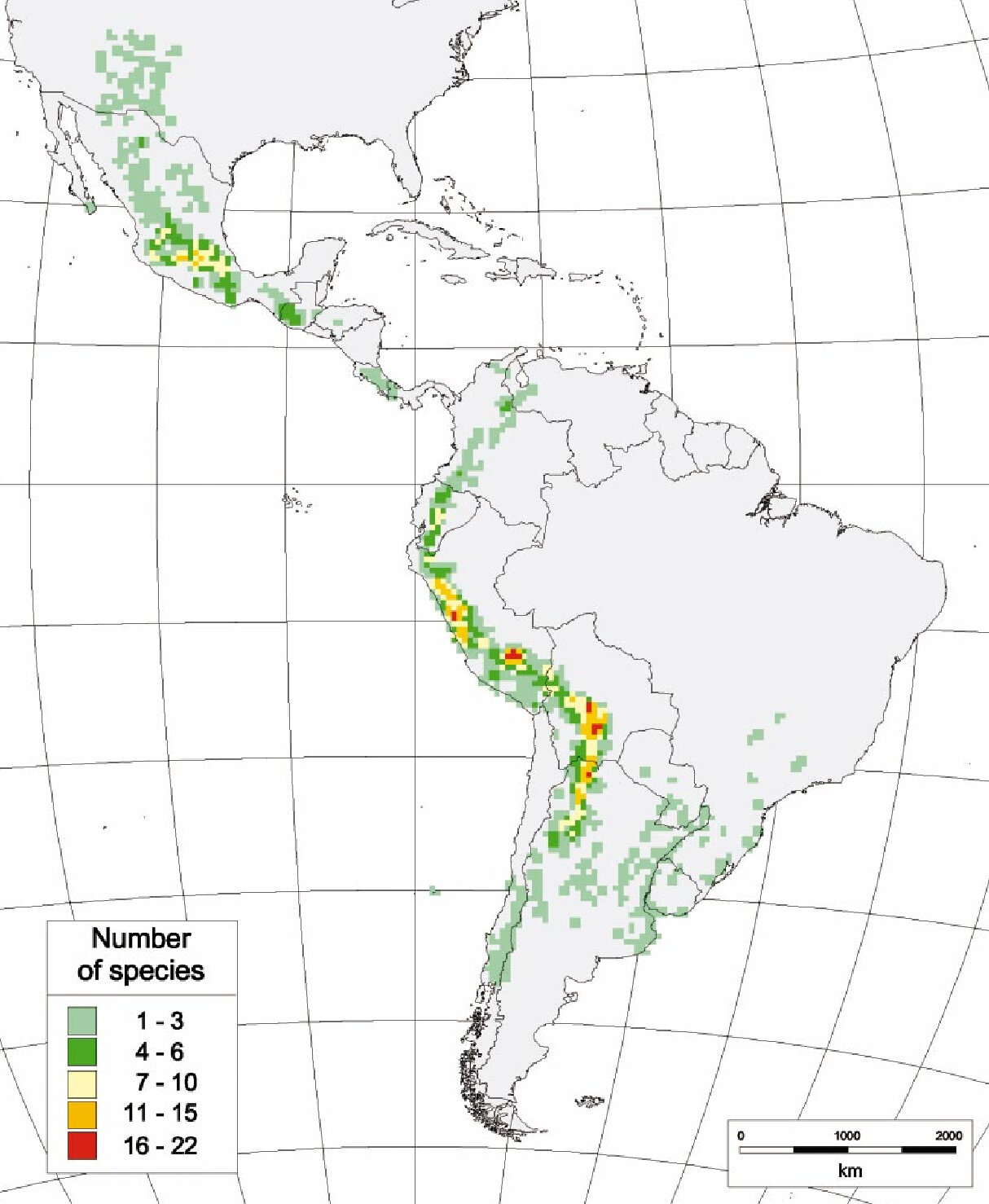
Current potato breeding efforts aim to improve yields, food processing qualities, resistance to abiotic stress (e.g. drought), and resistance to pathogens [5, 6]. In their search for traits of interest to be introgressed into S. tuberosum, breeders can rely on the large botanical diversity of potatoes encompassing 4 cultivated and 107 wild species [2], distributed throughout Latin America (Figure 1), from New Mexico to Patagonia [7, 8].
Being adapted to a broad array of environmental and climatic conditions, wild potatoes possess desirable agricultural traits, such as higher disease and drought resistance. Recent genomic studies in the potato clade (Solanum sect. Petota) indeed revealed the extraordinary diversity of wild potato germplasm available for potato improvement [9, 10].
S. chacoense: a resource for potato breeding…
My project deals with the reproductive biology of Solanum chacoense (Figure 2), a wild potato species whose genome was recently sequenced [11]. In breeding research, S. chacoense is studied for its high capacity to resist to a wide range of pathogens, such as potato virus Y [12, 13], bacteria causing potato wilt [14, 15] and common scab [16, 17], or fungi responsible for potato black scurf and blackleg [18], and powdery mildew [19].
Solanum chacoense has also a peculiar secondary metabolism producing high levels of specific glycoalkaloid compounds, such as leptines and leptinines [20], that are particulary efficient against the Colorado potato beetle [21]. This small herbivore insect (Figure 3) is a major potato pest that can completely destroy cultures by feeding on leaves. Effort is being made to better understand genes involved in those metabolic pathways [22, 23] and to introduce them into the cultivated potato [24, 25, 26, 27].
…and a model for plant reproduction research
Being a diploid species, S. chacoense is also studied as an alternative model to the tetraploid cultivated potato for molecular and cellular biology, especially in plant reproduction research. For instance, in our research institute, S. chacoense has been used in the Cappadocia lab to decipher the mechanisms underlying S-RNase-based gametophytic self-incompatibility [28, 29, 30, 31]. The Geitmann lab, now at McGill University, used S. chacoense in their research about cytomechanics of pollen tube growth [32, 33, 34].
The Matton lab uncovered various proteins involved in reproductive signal transduction pathways in S. chacoense, including RALF peptides [35, 36], MAP kinases (missing reference) and receptor-like kinases [37, 38, 39, 40]. Our lab also produced large-scale transcriptomic [41], proteomic [42] and secretomic [43] studies about S. chacoense reproduction.
My PhD project aims at understanding how pollen–pistil interactions, especially pollen tube guidance, are involved in the reproductive isolation of wild potatoes, i.e. the mechanisms by which they avoid interspecific hybridization.


Literature cited
- Spooner DM, McLean K, Ramsay G, Waugh R and Bryan GJ. (2005). A single domestication for potato based on multilocus amplified fragment length polymorphism genotyping. Proc. Natl. Acad. Sci. U. S. A., 102(41): 14694–9. DOI: 10.1073/pnas.0507400102
- Spooner DM, Ghislain M, Simon R, Jansky SH and Gavrilenko T. (2014). Systematics, diversity, genetics, and evolution of wild and cultivated potatoes. Bot. Rev., 80(4): 283–383. DOI: 10.1007/s12229-014-9146-y
- Birch PRJ, Bryan G, Fenton B, Gilroy EM, Hein I, Jones JT, … Toth IK. (2012). Crops that feed the world 8: Potato: are the trends of increased global production sustainable? Food Sec., 4(4): 477–508. DOI: 10.1007/s12571-012-0220-1
- Food and of the United Nations AO. (2016). FAO Statistics Division data for year 2016. URL: http://www.fao.org/faostat/en (accessed August 1, 2018).
- Halterman D, Guenthner J, Collinge S, Butler N and Douches D. (2015). Biotech potatoes in the 21st century: 20 years since the first biotech potato. Am. J. Potato Res., 93(1): 1–20. DOI: 10.1007/s12230-015-9485-1
- Hameed A, Zaidi SS-E-A, Shakir S and Mansoor S. (2018). Applications of new breeding technologies for potato improvement. Front. Plant Sci., 9: 925. DOI: 10.3389/fpls.2018.00925
- Hijmans RJ and Spooner DM. (2001). Geographic distribution of wild potato species. Am. J. Bot., 88(11): 2101–12.
- Hijmans RJ, Spooner DM, Salas AR, Guarino L and de la Cruz J. (2002). Atlas of wild potatoes. Rome, Italy: International Plant Genetic Resources Institute.
- Hardigan MA, Laimbeer FPE, Newton L, Crisovan E, Hamilton JP, Vaillancourt B, … Buell CR. (2017). Genome diversity of tuber-bearing Solanum uncovers complex evolutionary history and targets of domestication in the cultivated potato. Proc. Natl. Acad. Sci. U. S. A. DOI: 10.1073/pnas.1714380114
- Li Y, Colleoni C, Zhang J, Liang Q, Hu Y, Ruess H, … Huang B. (2018). Genomic analyses yield markers for identifying agronomically important genes in potato. Mol. Plant, 11(3): 473–84. DOI: 10.1016/j.molp.2018.01.009
- Leisner CP, Hamilton JP, Crisovan E, Manrique-Carpintero NC, Marand AP, Newton L, … Buell CR. (2018). Genome sequence of M6, a diploid inbred clone of the high-glycoalkaloid-producing tuber-bearing potato species Solanum chacoense, reveals residual heterozygosity. Plant J., 94(3): 562–70. DOI: 10.1111/tpj.13857
- Bravo-Almonacid F, Rudoy V, Welin B, Segretin ME, Bedogni MC, Stolowicz F, … Mentaberry A. (2012). Field testing, gene flow assessment and pre-commercial studies on transgenic Solanum tuberosum spp. tuberosum (cv. Spunta) selected for PVY resistance in Argentina. Transgenic Res., 21(5): 967–82. DOI: 10.1007/s11248-011-9584-9
- Duan H, Richael C and Rommens CM. (2012). Overexpression of the wild potato eIF4E-1 variant Eva1 elicits Potato virus Y resistance in plants silenced for native eIF4E-1. Transgenic Res., 21(5): 929–38. DOI: 10.1007/s11248-011-9576-9
- Bae J, Halterman D and Jansky S. (2008). Development of a molecular marker associated with Verticillium wilt resistance in diploid interspecific potato hybrids. Mol. Breed., 22(1): 61–9. DOI: 10.1007/s11032-008-9156-8
- Chen L, Guo X, Xie C, He L, Cai X, Tian L, … Liu J. (2013). Nuclear and cytoplasmic genome components of Solanum tuberosum + S. chacoense somatic hybrids and three SSR alleles related to bacterial wilt resistance. Theor. Appl. Genet., 126(7): 1861–72. DOI: 10.1007/s00122-013-2098-5
- Braun SR, Endelman JB, Haynes KG and Jansky SH. (2017). Quantitative trait loci for resistance to common scab and cold-induced sweetening in diploid potato. Plant Genome, 10(3). DOI: 10.3835/plantgenome2016.10.0110
- Jansky S, Douches D and Haynes K. (2017). Transmission of scab resistance to tetraploid potato via unilateral sexual polyploidization. Am. J. Potato Res., 95(3): 272–7. DOI: 10.1007/s12230-017-9628-7
- Almasia NI, Bazzini AA, Hopp HE and Vazquez-Rovere C. (2008). Overexpression of snakin-1 gene enhances resistance to Rhizoctonia solani and Erwinia carotovora in transgenic potato plants. Mol. Plant Pathol., 9(3): 329–38. DOI: 10.1111/j.1364-3703.2008.00469.x
- Faccio P, Vazquez-Rovere C, Hopp E, González G, Décima-Oneto C, Favret E, … Franzone P. (2011). Increased tolerance to wheat powdery mildew by heterologous constitutive expression of the Solanum chacoense. Czech J. Genet. Plant Breed., 47(Special issue): S135–41.
- Mweetwa AM, Hunter D, Poe R, Harich KC, Ginzberg I, Veilleux RE and Tokuhisa JG. (2012). Steroidal glycoalkaloids in Solanum chacoense. Phytochemistry, 75: 32–40. DOI: 10.1016/j.phytochem.2011.12.003
- Lorenzen JH, Balbyshev NF, Lafta AM, Casper H, Tian X and Sagredo B. (2001). Resistant potato selections contain leptine and inhibit development of the Colorado potato beetle (Coleoptera: Chrysomelidae). J. Econ. Entomol., 94(5): 1260–7.
- Manrique-Carpintero NC, Tokuhisa JG, Ginzberg I, Holliday JA and Veilleux RE. (2013). Sequence diversity in coding regions of candidate genes in the glycoalkaloid biosynthetic pathway of wild potato species. G3: Genes, Genomes, Genet., 3(9): 1467–79. DOI: 10.1534/g3.113.007146
- Manrique-Carpintero NC, Tokuhisa JG, Ginzberg I and Veilleux RE. (2014). Allelic variation in genes contributing to glycoalkaloid biosynthesis in a diploid interspecific population of potato. Theor. Appl. Genet., 127(2): 391–405. DOI: 10.1007/s00122-013-2226-2
- Cooper SG, Douches DS and Grafius EJ. (2009). Combining engineered resistance, avidin, and natural resistance derived from Solanum chacoense bitter to control Colorado potato beetle (Coleoptera: Chrysomelidae). J. Econ. Entomol., 102(3): 1270–80.
- Ginzberg I, Thippeswamy M, Fogelman E, Demirel U, Mweetwa AM, Tokuhisa J and Veilleux RE. (2012). Induction of potato steroidal glycoalkaloid biosynthetic pathway by overexpression of cDNA encoding primary metabolism HMG-CoA reductase and squalene synthase. Planta, 235(6): 1341–53. DOI: 10.1007/s00425-011-1578-6
- Molnár I, Besenyei E, Thieme R, Thieme T, Aurori A, Baricz A, … Rakosy-Tican E. (2017). Mismatch repair deficiency increases the transfer of antibiosis and antixenosis properties against Colorado potato beetle in somatic hybrids of Solanum tuberosum + S. chacoense. Pest Manag. Sci., 73(7): 1428–37. DOI: 10.1002/ps.4473
- Crossley MS, Schoville SD, Haagenson DM and Jansky SH. (2018). Plant resistance to Colorado potato beetle (Coleoptera: Chrysomelidae) in diploid F2 families derived from crosses between cultivated and wild potato. J. Econ. Entomol. DOI: 10.1093/jee/toy120
- Boivin N, Morse D and Cappadocia M. (2014). Degradation of S-RNase in compatible pollen tubes of Solanum chacoense inferred by immunogold labeling. J. Cell Sci., 127(Pt 19): 4123–7. DOI: 10.1242/jcs.154823
- Soulard J, Boivin N, Morse D and Cappadocia M. (2014). eEF1A is an S-RNase binding factor in self-incompatible Solanum chacoense. PLoS One, 9(2): e90206. DOI: 10.1371/journal.pone.0090206
- Soulard J, Qin X, Boivin N, Morse D and Cappadocia M. (2013). A new dual-specific incompatibility allele revealed by absence of glycosylation in the conserved C2 site of a Solanum chacoense S-RNase. J. Exp. Bot., 64(7): 1995–2003. DOI: 10.1093/jxb/ert059
- Liu B, Boivin N, Morse D and Cappadocia M. (2012). A time course of GFP expression and mRNA stability in pollen tubes following compatible and incompatible pollinations in Solanum chacoense. Sex. Plant Reprod., 25(3): 205–13. DOI: 10.1007/s00497-012-0192-5
- Aouar L, Chebli Y and Geitmann A. (2010). Morphogenesis of complex plant cell shapes: the mechanical role of crystalline cellulose in growing pollen tubes. Sex. Plant Reprod., 23(1): 15–27. DOI: 10.1007/s00497-009-0110-7
- Parre E and Geitmann A. (2005). Pectin and the role of the physical properties of the cell wall in pollen tube growth of Solanum chacoense. Planta, 220(4): 582–92. DOI: 10.1007/s00425-004-1368-5
- Parre E and Geitmann A. (2005). More than a leak sealant. The mechanical properties of callose in pollen tubes. Plant Physiol., 137(1): 274–86. DOI: 10.1104/pp.104.050773
- Chevalier E, Loubert-Hudon A and Matton DP. (2013). ScRALF3, a secreted RALF-like peptide involved in cell-cell communication between the sporophyte and the female gametophyte in a solanaceous species. Plant J., 73(6): 1019–33. DOI: 10.1111/tpj.12096
- Germain H, Chevalier E, Caron S and Matton DP. (2005). Characterization of five RALF-like genes from Solanum chacoense provides support for a developmental role in plants. Planta, 220(3): 447–54. DOI: 10.1007/s00425-004-1352-0
- Germain H, Gray-Mitsumune M, Houde J, Benhamman R, Sawasaki T, Endo Y and Matton DP. (2013). The Solanum chacoense ovary receptor kinase 11 (ScORK11) undergoes tissue-dependent transcriptional, translational and post-translational regulation. Plant Physiol. Biochem., 70: 261–8. DOI: 10.1016/j.plaphy.2013.05.036
- Germain H, Gray-Mitsumune M, Lafleur E and Matton DP. (2008). ScORK17, a transmembrane receptor-like kinase predominantly expressed in ovules is involved in seed development. Planta, 228(5): 851–62. DOI: 10.1007/s00425-008-0787-0
- Germain H, Houde J, Gray-Mitsumune M, Sawasaki T, Endo Y, Rivoal J and Matton DP. (2007). Characterization of ScORK28, a transmembrane functional protein receptor kinase predominantly expressed in ovaries from the wild potato species Solanum chacoense. FEBS Lett., 581(26): 5137–42. DOI: 10.1016/j.febslet.2007.10.001
- Germain H, Rudd S, Zotti C, Caron S, O’Brien M, Chantha S-C, … Matton DP. (2005). A 6374 unigene set corresponding to low abundance transcripts expressed following fertilization in Solanum chacoense Bitt, and characterization of 30 receptor-like kinases. Plant Mol. Biol., 59(3): 515–32. DOI: 10.1007/s11103-005-0536-8
- Tebbji F, Nantel A and Matton DP. (2010). Transcription profiling of fertilization and early seed development events in a solanaceous species using a 7.7 K cDNA microarray from Solanum chacoense ovules. BMC Plant Biol., 10: 174. DOI: 10.1186/1471-2229-10-174
- Vyetrogon K, Tebbji F, Olson DJH, Ross ARS and Matton DP. (2007). A comparative proteome and phosphoproteome analysis of differentially regulated proteins during fertilization in the self-incompatible species Solanum chacoense Bitt. Proteomics, 7(2): 232–47. DOI: 10.1002/pmic.200600399
- Liu Y, Joly V, Dorion S, Rivoal J and Matton DP. (2015). The plant ovule secretome: a different view toward pollen–pistil interactions. J. Proteome Res., 14(11): 4763–75. DOI: 10.1021/acs.jproteome.5b00618